Origins of Lager Yeast

The domestication of barley coincides with the first year-round settlements of early humans, who were previously nomadic. This event occurred in the Fertile Crescent — the region surrounding the Tigris and Euphrates rivers, and sometimes extended to include the Nile Delta — 6,000 years ago. In this same time period, brewing was invented, as shown by evidence of brewing in ancient Sumer by this period. (Sumer was mostly located between the Tigris and Euphrates, in what is currently south-eastern Iraq.) Subsequently, numerous plants and animals have been domesticated by human populations all over the world for agricultural, industrial and other purposes. And, of course, brewing spread to every human culture thousands of years ago.
It takes more than barley to make beer, however. Scientists have made substantial progress in understanding the domestication of most important macroscopic agricultural species. However, it has only been recently that they have begun to make real progress towards understanding the domestication of microscopic species. This includes the yeast strains used in brewing. The advent of DNA sequencing technology, and particularly the newer methods of obtaining large amounts of sequence quickly and cheaply, have driven this progress.
Finding the wild progenitor (or a closely related species) of a domesticated organism helps scientists determine how that species was domesticated. In the classic example, scientists in the 1930s first recognized that wild species of teosinte (now classified as subspecies and named Zea mays parviglumis and Zea mays mexicana), found in Mexico and Central America, were related to domestic corn (maize, Zea mays). Later genetic studies revealed that the major differences between the grass teosinte and large-eared corn were due to only five genes. Modern sequence data shows that the modern corn genome is mostly derived from Zea mays parviglumis, with a small amount of sequence coming from hybridizing with Zea mays mexicana. Furthermore, estimates of the divergence time between teosinte and corn taken from comparative DNA sequence data mesh closely with the archaeological evidence that this happened roughly 9,000 years ago.
In addition, if a wild population of the ancestral species (or a close relative) still exists, this can be a source of genetic material to potentially improve the domesticated species. (During domestication, many potentially beneficial genetic variants from the parent stock can be lost.) To continue with the corn example, in 1977, scientists found a new species of teosinte (Zea diploperennis), which turned out to harbor several genes that conferred resistance to various viral diseases of corn. Using genes from this species, modern corn varieties were improved.
Recently, a group of scientists have reported finding a species of yeast that is very likely one of the ancestors of lager yeast. In order to appreciate the importance of this, it pays to understand a little of what is currently known about the origin of brewing strains of yeast.
Saccharomyces
Except for a few experimental beers brewed with Brettanomyces, beer is brewed with yeast from the genus Saccharomyces as the primary fermenting organism. Indeed, in the vast majority of brewery fermentations, other microbial species present are contaminants. Sour beers — in which some combination of Brettanomyces (wild yeast), Lactobacillus (bacteria) and Pediococcus (bacteria) may also be used — are the exception.
Saccharomyces is a genus of fungi that contains many species. Microbial taxonomy is continually in flux — with multiple former species being lumped together into one, former single species being split into two or more and new species being found and named all the time. So, citing an exact number would involve a number of assumptions. There are over twenty species common enough to have their own names. Saccharomyces species involved in brewing and winemaking include S. cerevisiae (ale and wine yeast), S. pastorianus (lager yeast), S. bayanus (used in some wine fermentations), S. uvarum (a contaminant that can grow at low temperatures; used to be considered a substrain of bayanus). The wild species S. paradoxus is the closest known species to S. cerevisiae.
Ale Yeast
Ale yeast (S. cerevisiae) is utilized by humans in baking leavened breads and conducting fermentations of ales, wines, saké, mead and cider. (Some wines are fermented with strains of S. bayanus.) Ale fermentations are generally conducted in the temperature range of 65–72 °F (18–22 °C). Ale yeast grows better at temperatures higher than this. However, at higher temperatures most ale strains would produce an ale that was too estery and might contain potentially harmful levels of fusel oils. In winery fermentations of red wine, fermentation temperatures up to 90 °F (32 °C) are not uncommon. Among Saccharomyces strains, cerevisiae is called thermotolerant.
S. cerevisiae is found in bakeries, breweries, wineries and in the wild. Specifically, in North America, S. cerevisiae is frequently found in the sugar-rich exudates of oak trees. Until recently, biologists were unsure if S. cerevisiae found in the wild was free-living wild population, or the result of continuing contamination from human sources. Recent genetic analysis has shown that wild populations of S. cerevisiae contain much more genetic diversity than domesticated strains and wild populations harbor alleles that are not present in domesticated strains. The implication is that S. cerevisiae does live in the wild, apparently with little interbreeding with domesticated strains.
S. cerevisiae is a diploid species, meaning its genome consists of two of each of its chromosomes. (Humans are also diploid, we inherit one set of chromosomes from our father and another set from our mother.)
“Other” Yeasts
Two other yeasts play ancillary roles in the story of lager yeast — Saccharomyces uvarum and S. bayanus. S. uvarum is a diploid yeast that is sometimes found as a contaminant in brewery or winery fermentations. S. uvarum is more tolerant of cold temperatures than S. cerevisiae (its cryotolerant, in the lingo). As a relative of S. cerevisiae, S. uvarum has a similar genome, but there is no evidence of recent hybridizations. (The level of sequence divergence between uvarum and cerevisiae is relatively constant across their whole genome.) Like S. cerevisiae, S. uvarum is found in the wild, in Europe and elsewhere.
S. bayanus is sometimes used in winery fermentations. Like both cerevisiae and uvarum, it is diploid. However, the pattern of sequence similarity between these species suggest that S. bayanus contains a mixture of genes recently derived from S. uvarum and an unidentified yeast, along with a minority of sequences from S. cerevisiae. S. bayanus is not found in the wild.
Lager Yeast
Lager yeast (Saccharomyces pastorianus) is used by brewers in the production of lager beer. Typical lager fermentations range from 48–55 °F (8.9–13 °C). Scientists call S. pastorianus cryotolerant, just like S. uvarum.
About 20 years ago, scientists discovered that lager yeast was a hybrid organism. The genome of S. pastorianus was found to be tetraploid (contains four copies of each chromosome, instead of two like the other yeasts we’ve discussed). The S. pastorianus genome was also known to contain sequences from S. cerevisiae and one other species of Saccharomyces. The other species was alternately hypothesized to be S. bayanus, S. uvarum (then called S bayanus var. uvarum), a combination of these two or an undiscovered cryotolerant species of Sacchar-omyces. Later work strongly suggested that S. pastorianus was a hybrid of cerevisiae and a yet-to-be-identified yeast.
The Discovery
Lager yeast is only found in breweries. It is not found in the wild in Europe — where lager brewing originated — or elsewhere. Likewise, searches for the cryotolerant component strain of the S. pastorianus hybrid species in Europe failed to uncover a match. So, it was somewhat surprising when researchers found the missing piece of the lager puzzle in Patagonia, a region in South America that overlaps part of Argentina and Chile.
The researchers, which included scientists from Argentina and the United States, discovered two strains of yeast living on Southern Beech trees (Nothofagus) in Patagonia. They found two different cryotolerant species, one associated with two different species of beech (N. antartica and N. pumilio) and one associated with a third species of beech (N. dombeyi). The two species did not appear to be interbreeding. The species associated with N. dombeyi was found to have sequences very similar to S. uvarum. The species associated with the other trees was a previously uncategorized species. When the researchers compared the genome of this new species to the genome of S. pastorianus, they found that its genome was highly similar to half of the genome of S. pastorianus — the missing species of yeast had been found. Because the new yeast also showed some sequence similarity to S. bayanus (which I’ll explain in a moment), they proposed that the new species be called S. eubayanus.
The Big Picture
If you take all the information that is known — including a few things not explained here — the whole story goes something like this. Somehow — and this has yet to be determined — a wild yeast from Patagonia contaminated a European brewery in the 15th Century. (Europe traded with South America at the time, so items made of beechwood carrying S. eubayanus may have shuttled the yeast there.) This diploid yeast (S. eubayanus) hybridized with an ale strain of S. cerevisiae (also diploid), producing the tetraploid ancestor of modern lager yeasts. This combination of a proven beer fermenter (cerevisiae) and a species that thrived in colder climates was selected accidentally as brewers cold-aged their beer.
Early pastorianus is thought to have contained roughly 50% eubayanus sequences and 50% cerevisiae sequences. However, new genomic sequencing data shows that many of the genes originally contributed by S. cerevisiae have been deleted, while almost all of the eubayanus sequences remain. In short, you can think of present day S. pastorianus as containing a full set of chromosomes from S. eubayanus and a set of “shrunken” chromosomes from S. cerevisiae. (This pattern of sequence lost after hybridization is common. The basic idea is that, following hybridization, duplicate genes eventually become deleted and are lost because the remaining gene “takes up the slack.”)
As S. pastorianus was domesticated, other genetic changes occurred as well. For example, a bit of one cerevisiae chromosome, that contained multiple copies of the gene maltase, got copied to the end of one of the pastorianus chromosomes. (The gene maltase produces the enzyme that degrades maltose, the main wort sugar.)
For brewers, that’s the most interesting bit, but for yeast biologists, the story has a final twist. Remember S. bayanus? Some of its sequences were similar enough to S. eubayanus to inspire its name. As it turns out, S. bayanus is the result of multiple hybridization events and contains sequences from S. cerevisiae, S. uvarum and S. pastorianus. However, unlike S. pastorianus — which contains a mix of chromosomes from its parent species — S. bayanus chromosomes themselves are a patchwork of sequences from the three species. (This indicates that a lot of genetic recombination has occurred in S. bayanus. In contrast, except for the “displaced” cerevisiae sequence tacked onto one of the pastorianus chromosomes, little recombination between cerevisiae and eubayanus chromosomes has occurred in S. pastorianus.) Like S. pastorianus, S. bayanus does not occur in the wild.
What Does This Mean?
What does this mean to us as brewers? In terms of how we handle lager yeast and conduct lager fermentations, very little. Nothing in the new information obviously suggests that changes are needed in any brewing practice. The value of the study will come when researchers begin to survey the genetic diversity found in wild populations of S. eubayanus. It is possible — likely even — that some alleles found in nature could be moved into domesticated S. pastorianus, either by hybridization or (more likely) genetic engineering, to improve lager yeasts.
Conclusion
So, lager yeast (S. pastorianus) arose in a European brewery via chance hybridization. The next time you brew a lager, you can take some vicarious pride in knowing that, without brewers, it would not exist.
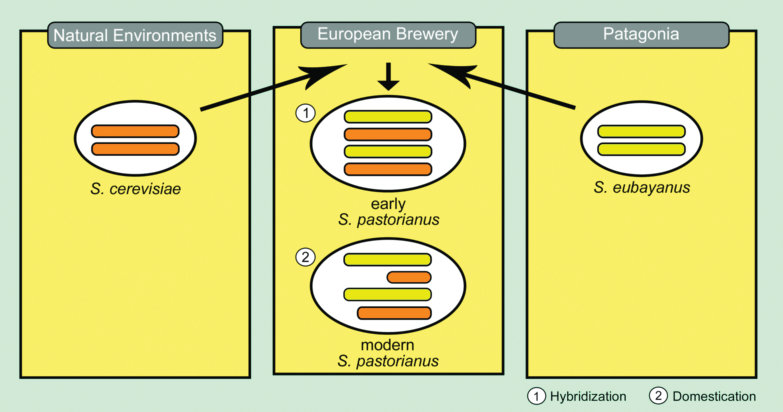
A diagram showing the basics of the origin of lager yeast. The orange and yellow rods represent chromosomes. S. cerevisiae was domesticated from the wild in ancient times. In the 15th Century, wild S. eubayanus from Patagonia ended up in a European brewery and hybridized with a domesticated strain of S. cerevisiae. Following hybridization, sequences originating in the cerevisiae genome were lost.


